|
Resources |
Bioinformatics |
|
|
Some
Bioinformatics:
Find restriction sites and fragment lenghtsnote: Use the program EMBOSS restrict.
More...
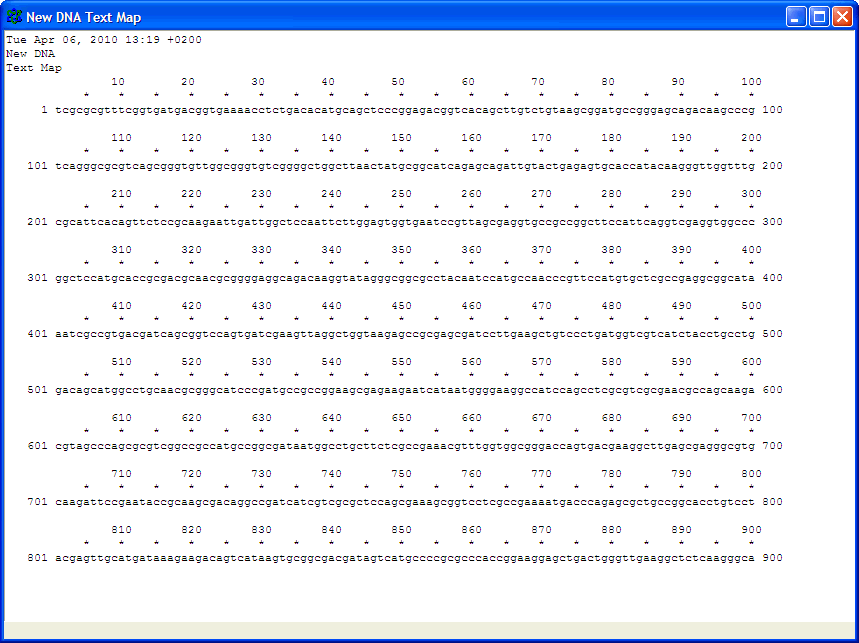
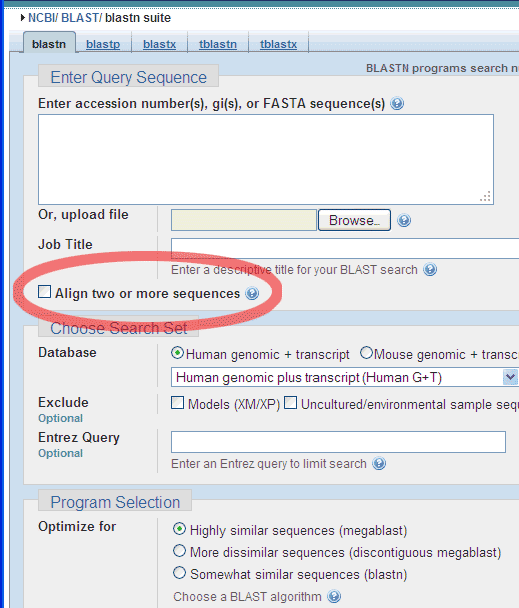
Number nucleotides in DNA sequences:For example: use ApE to number the nucleotides in this sequence: Use the Text Map feature and if you would leave this default settings: You would get this: Alignment of two given sequencesFor the alignment of two given sequences you can use the BLAST program bl2seq (Blast Two Sequences), for example to find out where a primer exactly anneales on a particular template.
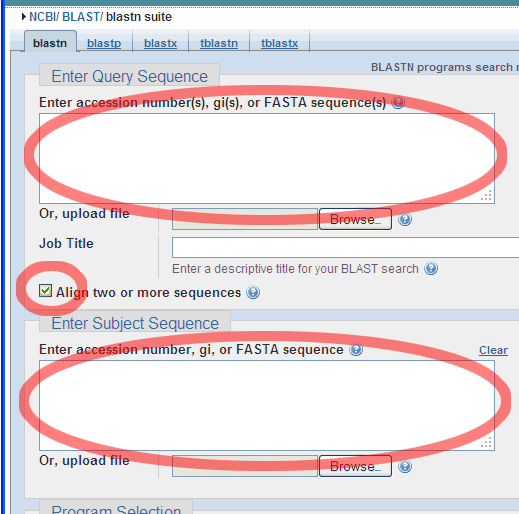
Choose the Align two or more sequences option:
And enter a Query Sequence (e.g. the smaller of the two Standard GeneTech
primers),
..like this:
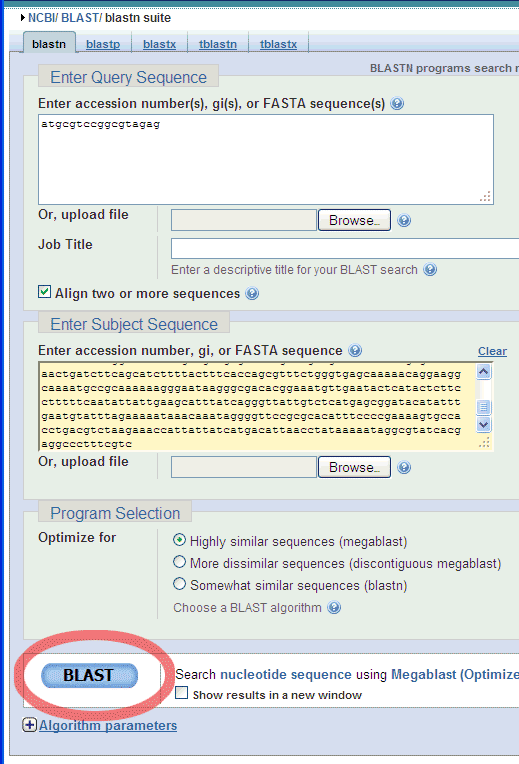
Then, scrol down to click the BLAST button:
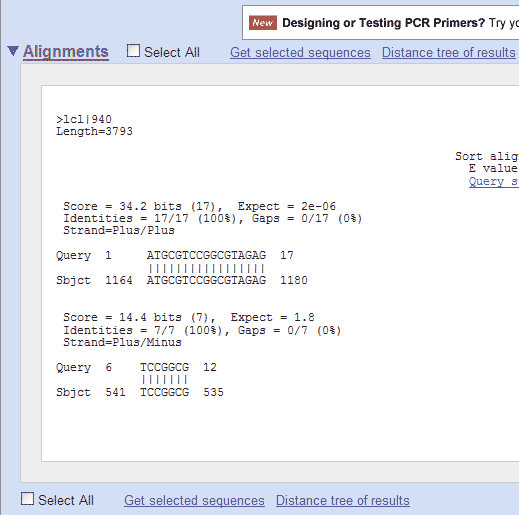
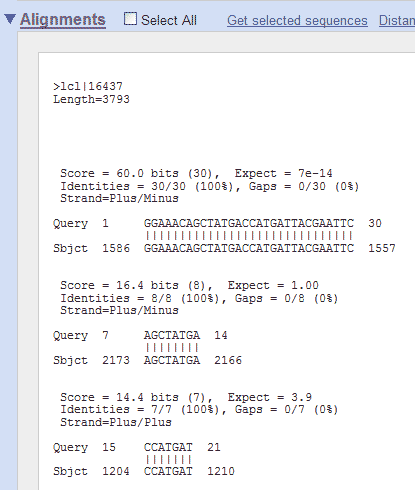
The resulting output screen shows the position in the pGT4 sequence, where homology with the primer is found:
So, in this example, the smaller (17 nucleotides long) Standard GeneTech primer has perfect homology (is identical to) with the pGT4 sequence from nucleotide position 1164 to 1180. If you would do the same for the longer (30 nucleotides long) Standard GeneTech primer, you would get this:
It shows you, that the longer primer is
identical to the pGT4 DNA sequence from position 1586 to 1557, which
means it is in the reverse orientation ("it anneals to the
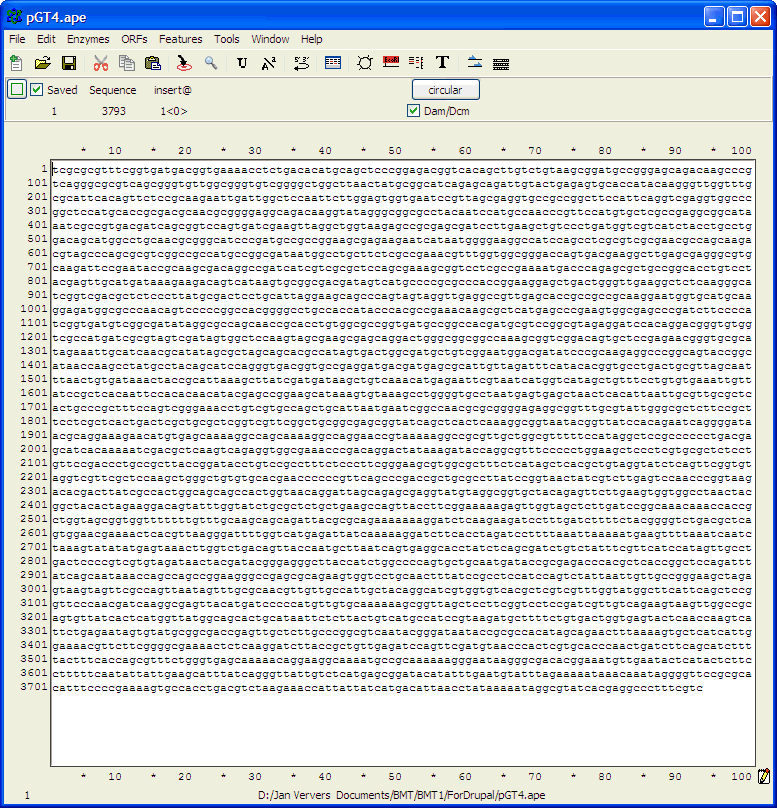
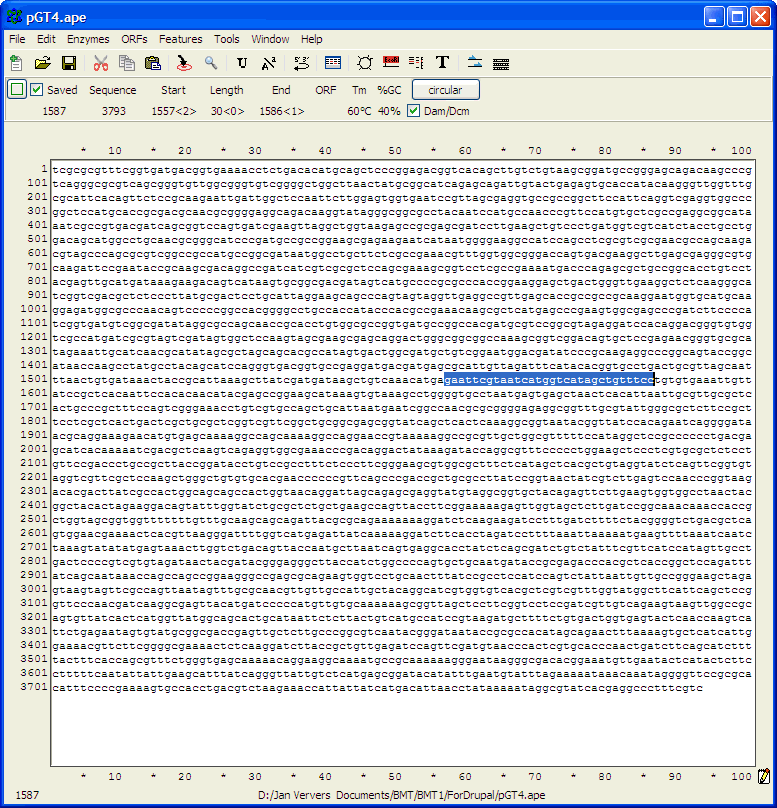
complementary strand and points backwards"). Finding primer binding sites using ApEIn this example, the ApE program is used to find the binding site on pGT4 DNA for primer ggaaacagctatgaccatgattacgaattc. Open ApE and paste the pGT4 sequence in the sequence field (or open the pGT4.ape file, if made before..). Make sure that after pasting a plasmid seq into ApE, the linear/circular button is set to circular!!
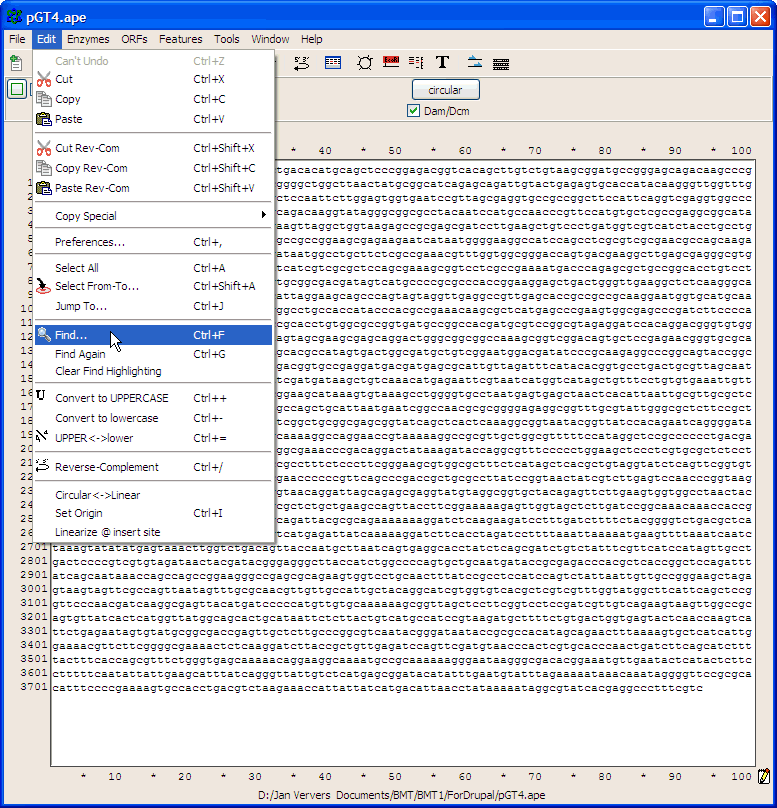
Go to Edit>Find (or use Ctrl+F)
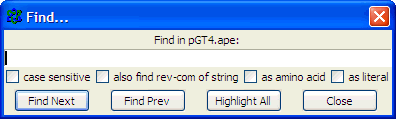
Paste the primer sequence
and check also find rev-com of string (the primer may be identical to (a part of) the opposite pGT4 DNA strand..)
and click the Find Next button to get this:
As seen by eye, the sequence in blue appears to be the
reverse-complement of the primer sequence.
Tm of PCR primersFor an accurate calculation of the Tm of PCR primers you could use the Oligonucleotide Properties Calculator at http://biotools.nubic.northwestern.edu/OligoCalc.html/.
about ORF FinderThe
ORF
Finder (Open Reading Frame Finder) tool finds all open reading
frames in a given sequence.
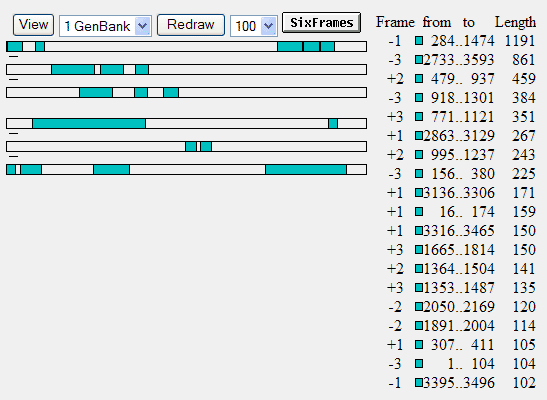
and click the OrfFind button (top left above the field) you would get this:
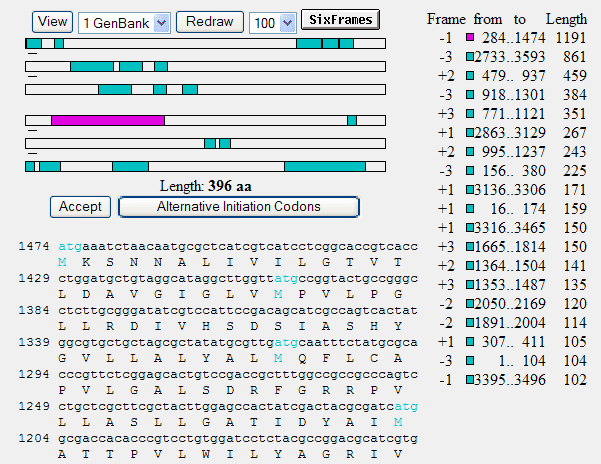
where ORFs are shown by green bars. The longest one runs from nucleotide position 284 to 1474. It's in the -1 (minus one) frame, so it starts at 1474 and stops after 284. This is the tetracyclin resistance gene of pGT4. If you click on that green bar (or the green square before 284), you get more detailed info about that reading frame:
[optinal] Try this:
|