























 |
 |
 |
|
 |
 |
||
 |
 |
||
 |
 |
||
 |
 |
||
 |
 |
||
 |
 |
||
 |
 |
||
 |
 |
||
 |
 |
||
 |
 |
||
 |
 |
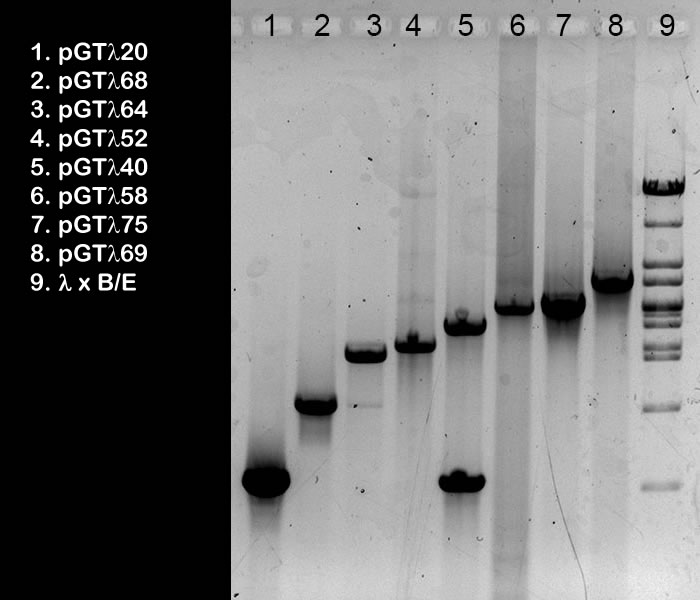
| This is a black-and-white
photograph of an agarose gel after elctrophoresis. In lanes 1-8, 5 ml samples from pooled PCR products of pGTλ clones were run. The gel was done (by wet lab groups A students) to check the quality and quantity of the final PCR products preps. They would later be used as template for probe labeling for the Southern hybrididsation step. The list on the left shows the clones on which the PCR was done. The insert of these clones were amplified using Standard GeneTech primers. Inserts can be easily recognised (only when you know exactly which fragments are visible in size marker lane 9! Realise that this sample was not heated prior to loading, so this band (K) is the cos-site containing 9035 fragment): The 1120 bp insert is visible in lane 1 (A), 1868 in lane 2 (B), 2564 in lane 3 (C), 2752 in lane 4 (D), 3240 in lane 5 (E), 3758 or 3775 in lane 6 (F), 3758 or 3775 in lane 7 (G), and 4669 in lane 8 (H). Purified product 3240 is contaminated with 1120 (I); this must be due to a mistake during pooling the PCR rreaction mixture. In lane 3 there’s a little contamination with 1868 (J), which could well be caused by overflow from slot 2 during gel loading.. If you would like to
estimate the concentration of the PCR products prep, you could
do like this:
|