About calculation of fragment lengths by the EMBOSS program:
An example:
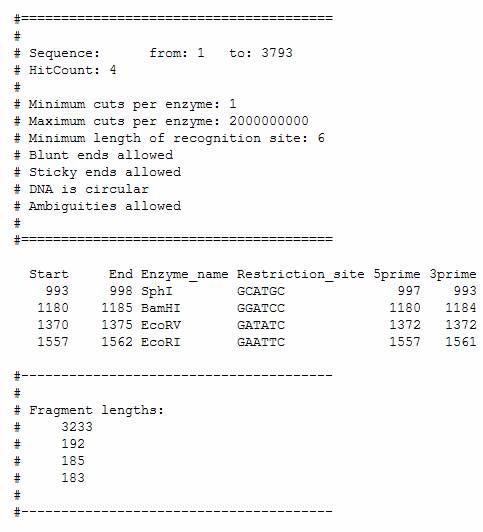
When EMBOSS calculates the lengths of the DNA fragments in a BamHI/EcoRI/EcoRV/SphI digest of plasmid pGT4, the program gives the following output:

In the 5prime column, EMBOSS shows the position of the last nucleotide before the cut in the upper strand (the one shown in a sequence text, from 5’ (left) to 3’ (right)). The 3prime column shows the position of the last nucleotide before the cut in the lower strand (the one usually not shown in a sequence text).
In the daily lab practice you would probably say:
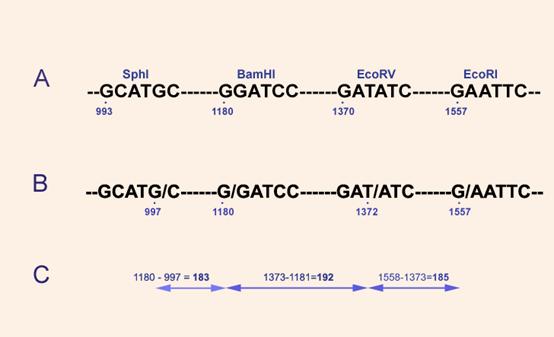
“There is an Sph site at 993, a Bam site at
1180, an Eco site at 1557, and an EcoRV site at 1370 [A in
figure below] .
This will give 1180 – 993 = 187, 1370 – 1180 = 190, and 1557 – 1370 =
187 for the small fragment sizes”.
This is not accurate.
Programs like EMBOSS look at the exact cut positions (B in
figure below)
and then calculate the lengths (C
in figure below).