Example:
How to use the EMBOSS programs to find restriction sites and fragment lengths in plasmid vector pGT4.
Allow circular DNA
Show fragment lengths
Find single cutters
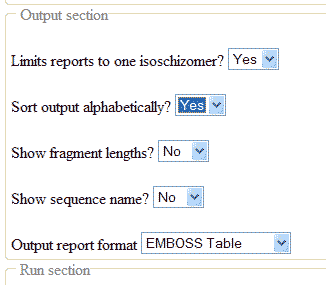
Sort output alphabetically
Go to http://emboss.bioinformatics.nl/
In the left pane: scroll down till the nucleic Restriction group

and choose restrict:
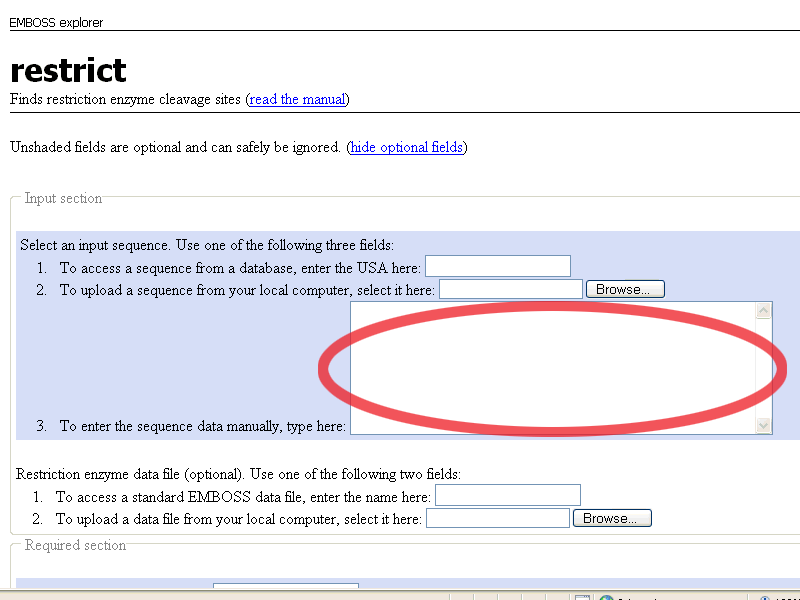
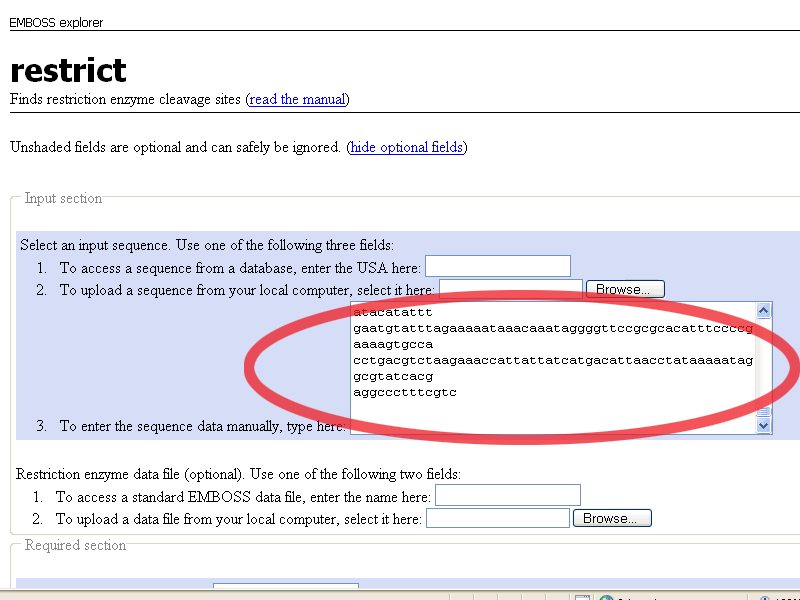
When the restrict page is open, paste the pGT4 DNA sequence in the To enter the sequence manually field
This is what you get:
Scroll down, and leave all options default except
Comma separated enzyme
list where you should fill in
bamhi,ecori
(case-insensitive, but use a comma and no spaces between the enzyme names..)
Allow circular DNA?
where you should indicate
yes (since pGT4 is circular DNA)
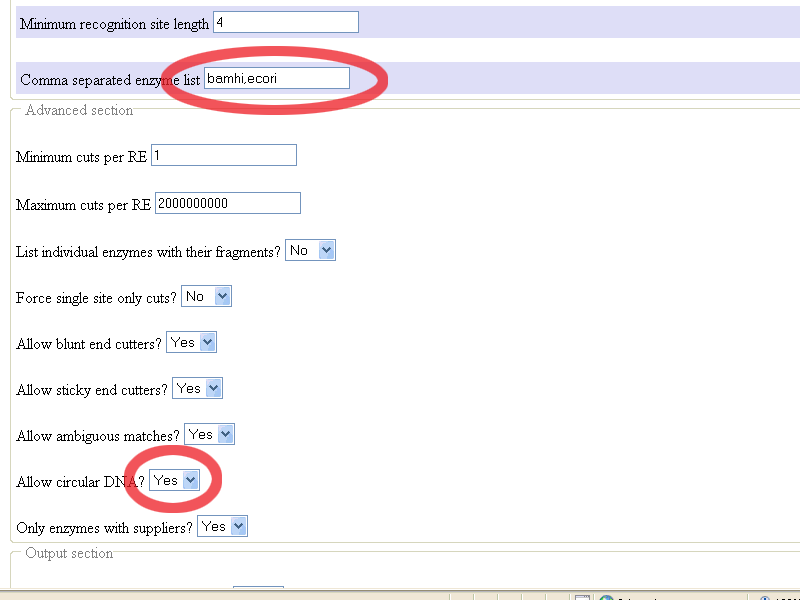
After changing
the default Show fragment lengths? No to Yes,
click the Run restrict button.

This is what you see first:
Don't worry, just scroll down untill you see this:
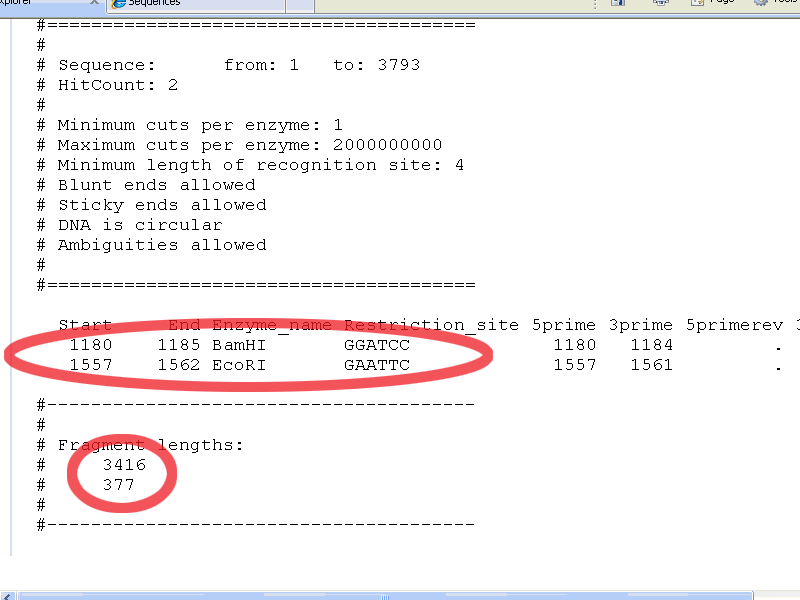
which shows you, that pGT4 (3793 basepairs) has a (apparently unique) BamHI restriction site at position 1180 (it is GGATCC from position 1180 to 1185) and an (also unique) EcoRI restriction site at position 1557 (it is GAATTC from position 1557 to 1562).
A digestion with
both enzymes gives fragments of 3416 and 377 basepairs.
(note: Sometimes the EMBOSS restrict output gives unexpected results.
More..)
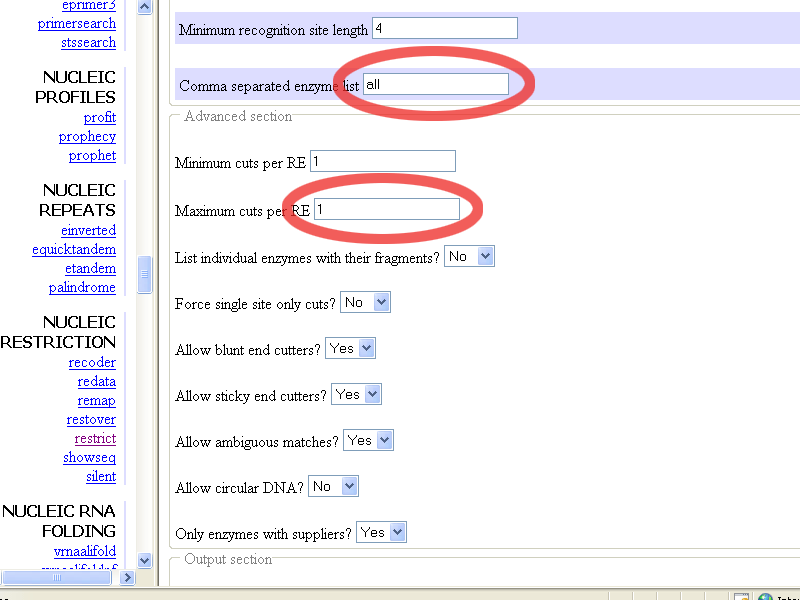
If you would
like to find all
single-cutters for pGT4 (restriction enzymes that have only
one recognition site in pGT4) do the following:
leave or fill-in all in the Comma separated enzym list, and set the
Minimum and the Maximum cuts per RE to 1, to get a complete list with unique
sites in pGT4, like this:
To get a list with the single-cutters in alphabetical order, set the option in the Output section, like this: