|
Bioinformatics
Sequence notation
Lambda DNA
pGT4
Restriction enzymes
pGTλclones
Sequences
Links
|
About Lambda DNA
The sequence of
bacteriophage Lambda DNA.
Lambda
digests gel
About samples containing Lambda-DNA:
Lambda is a medium size E.coli bacteriophage. The DNA molecule of
48502 basepairs is linear
and except for the extreme ends double-stranded. At each end the 5' strand overhangs
the 3' strand by 12 bases. The sequences of the ends are complementary. At ambient
temperatures, in a solution containing purified Lambda-DNA these so-called 'cos
ends' may pair and form the so-called 'cos-site'. As a consequence, the
DNA is (partly) circularised or have formed concatemers.
In a purified- DNA
solution the cos-site can be destroyed by heating 10 minutes at 65-70 °C. Immediate
cooling in ice-water prevents reformation of the "cos-site".
note:
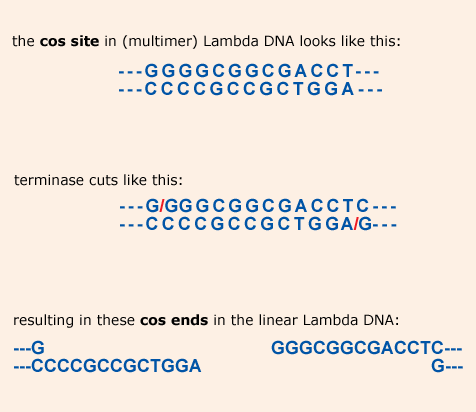
The 12 nucleotide 5’ overhangs at the cos-ends of the linear Lambda DNA
are the result of a cut by the enzyme terminase. This enzyme is
encoded by Lambda itself and acts like a restriction enzyme during the
replication of the phage DNA. It is an endonuclease specific for the
cos-site in multimeric phage DNA. The ends of the resulting monomeric
DNA (called cos ends) are similar to the sticky (or cohesive)
ends produced by common restriction enzymes. Because they have long
5’ overhangs (12 nucleotides) cos ends are much more sticky than the
cohesive ends generated by restriction enzymes which have generally an
overhang of 4 or 2 nucleotides.
Since cos ends have complementary
overhangs they are compatible for efficient ligation.
The ends in detail:
After electrophoresis, the gel band pattern of a Lambda DNA digest
may show an extra band, due to the joining (and forming the
cos-site) of the two end fragments. If this happens (it will always, to some
extent..), the bands containing the separate end fragments will be
present in an amount lower than calculated..
Example:
| |
|
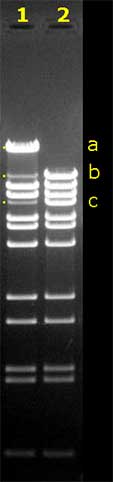 |
The gel on the left shows the gel electrophoresis result
of two BstE II digests of Lambda DNA.
Before pipetting the samples into the slots, the digest for
lane 2 was heated for 10 minutes at 70°C, to destroy the
cos-site.When the digest is not heated before the gel
run (lane 1), a number of b and c fragments have joined
to form band a, which is in size the sum of b
and c.
It is very likely that a number of the Lambda DNA
molecules was circular (because of the cos-site formed)
already before the digestion.
When you use EMBOSS>Restrict to find out
about the fragment sizes, you will not find band a.
You will also find out, that b and c are
end fragments.
When you use EMBOSS>restrict to "do" the BstE II digestion
with the option Allow
circular DNA? > YES, you will not find
fragments b and c in the output list, but
fragment a instead.
When you use ApE to find the BstE II fragment lengths,
it will depend on state of the linear/circular button
(top right in the ApE window) whether you'll find a,
or b and c.
|
note:
Restriction enzymes often generate "cohesive" or "sticky" ends. Those are ends
with short, mostly 4 nucleotides long, single-stranded overhangs. Because the
overhangs are much shorter than the Lambda cos-ends, annealing of those ends
is much less stable. After gel electrophoresis, you'll never observe additional
DNA bands, formed by fragments joined by restriction enzyme generated "sticky"
ends.
Map
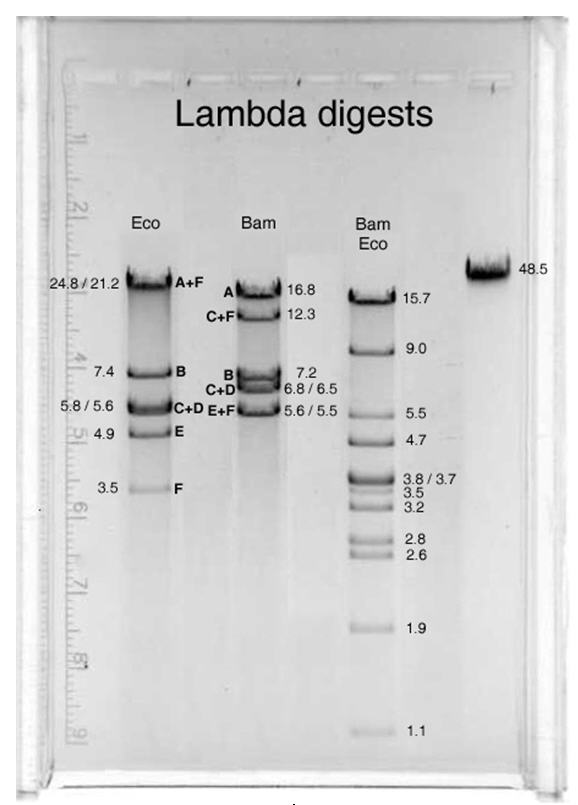
This is a BamHI-EcoRI restriction map of Lambda DNA:

(read it like this: (e.g.) "the 4878 bp long EcoRI fragment is between
the first and the second EcoRI site, and in the Simple Cloning Lab it is
called Eco E" ).
The staggered ends of the black bar indicate the single-stranded cos ends of
the Lambda DNA, which can anneal to form the cos site (BamC, BamF,
EcoA and EcoF are end fragments).
The exact positions
of the BamHI and EcoRI (and other) restriction sites can be found in the table
below.
Restriction sites list
An output file from EMBOSS>restrict for restriction enzyme sites in
Lambda DNA can be found
here. The list
shows all enzymes with a 6 basepairs recognition site ("6cutters") in
alphabetical order.
|




