|
Resources |
Sequence notation |
|
|
Sequence notation
About sequence notation:Standard notation of DNA sequences is from 5’ to 3’. So,
primer sequence atgcgtccggcgtagag means 5’
atgcgtccggcgtagag 3’
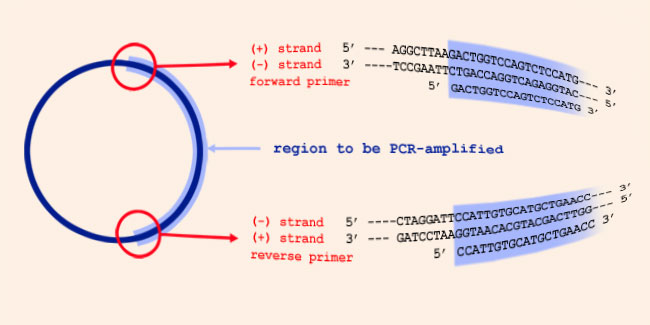
About primers and their direction in PCR:
The orientation of primers means: the direction in which the elongation
of the primer in DNA synthesis occurs. Since DNA synthesis is always from
5' to 3' , the 3' ends of a PCR primer set point towards each other, when
they are annealed to their template strand, and the primers anneal on
opposite strands of the PCR template. In the picture above, the forward
primer anneals to the template (-) strand, and is identical to (a part of)
the template (+) strand. And the reverse primer anneals to the template
(+) strand, and is identical to (a part of) the template (-) strand.
Forward, reverse, (+) and (-) refer to transcription of genes: the (+) DNA
strand has the same orientation as a messenger RNA, transcribed from the
DNA. So the forward orientation is: pointing towards the end of a gene. Restrictrion enzymes recognition sites nomenclatureRecognition sequences representations use the standard abbreviations to represent ambiguity:
R = G or A
Recognition sequences are written from 5' to 3', only one strand being given. If the point of cleavage has been determined, the precise site may be marked with ^. For enzymes such as HgaI, MboII etc., which cleave away from their recognition sequence the cleavage sites are indicated in parentheses. For example HgaI GACGC (5/10) indicates cleavage as follows:
Typically, the recognition sequences are oriented so that the cleavage sites lie on their 3' side. note: Reverse complementOften we need to obtain the complementary strand of a DNA sequence. As DNA is antiparallel, we really need the reverse complement sequence to keep our 5' and 3' ends properly oriented. While this is easy to do manually with short sequences, for longer sequences computer programs (e.g. EMBOSS revseq) are easier. Original
Sequence 5'ATGCAGGGGAAACATGATTCAGGAC
3' (Complement Pairs with Original Sequence, antiparallel)
(= Complement sequence written 5' to 3') In a palindromic DNA sequence, the sequence is the same when one
DNA strand is read left to right and the other strand is read right to
left. So, the reverse complement of the palindromic sequence is the same
sequence.. |