Interaction network for RSM_4_17.35
| Layout: | |
| Zoom | |
|
|
|
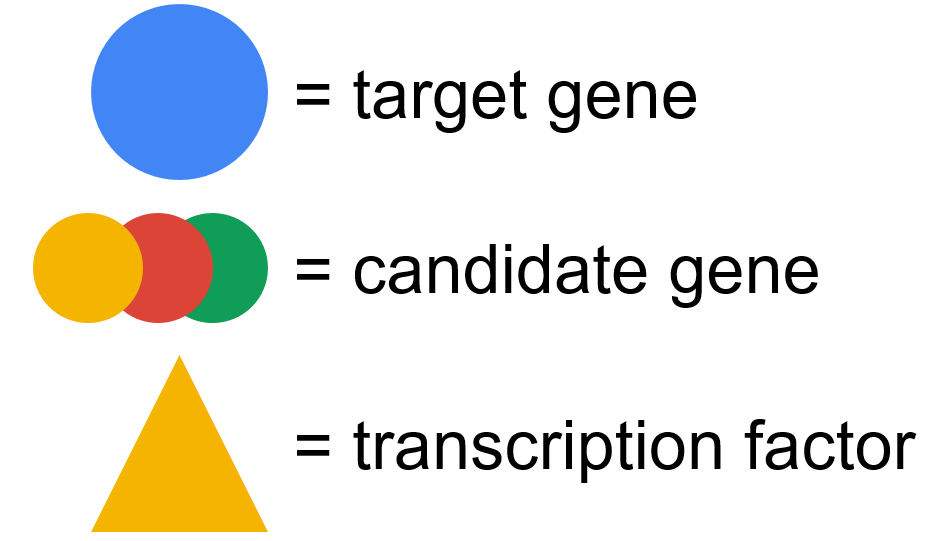
Please select a peak in the plot
| Layout: | |
| Zoom | |
|
|
|

| Trait (count=17) | Type | Name | LOD score | Position | Description | ||
|---|---|---|---|---|---|---|---|
| AT4G36760 | gene | APP1 | 14.57 | 4:17326537 | Arabidopsis aminopeptidase P1 | ||
| AT4G37320 | gene | CYP81D5 | 11.78 | 4:17559378 | member of CYP81D | ||
| AT4G37210 | gene | 11.65 | 4:17512254 | Tetratricopeptide repeat (TPR)-like superfamily protein%3B FUNCTIONS IN: binding%3B INVOLVED IN: biological_process unknown%3B LOCATED IN: cellular_component unknown%3B EXPRESSED IN: 23 plant structures%3B EXPRESSED DURING: 14 growth stages%3B CONTAINS InterPro DOMAIN/s: Tetratricopeptide TPR-1 (InterPro:IPR001440)%2C Tetratricopeptide-like helical (InterPro:IPR011990)%2C Tetratricopeptide repeat-containing (InterPro:IPR013026)%2C Tetratricopeptide repeat (InterPro:IPR019734)%3B Has 30201 Blast hits to 17322 proteins in 780 species: Archae - 12%3B Bacteria - 1396%3B Metazoa - 17338%3B Fungi - 3422%3B Plants - 5037%3B Viruses - 0%3B Other Eukaryotes - 2996 (source: NCBI BLink). | |||
| AT4G37150 | gene | MES9 | 10.54 | 4:17492871 | Encodes a protein shown to have carboxylesterase activity%2C methyl salicylate esterase activity%2C methyl jasmonate esterase activity%2C and methyl IAA esterase activity in vitro. MES9 appears to be involved in MeSA hydrolysis in planta. Expression of MES9 can restore systemic acquired resistance in SAR-deficient tobacco plants. This protein does not act on MeGA4%2C or MEGA9 in vitro. | ||
| AT4G37370 | gene | CYP81D8 | 8.76 | 4:17569740 | member of CYP81D | ||
| AT4G37440 | gene | 8.69 | 4:17601185 | unknown protein%3B BEST Arabidopsis thaliana protein match is: unknown protein (TAIR:AT3G50040.1)%3B Has 220 Blast hits to 205 proteins in 55 species: Archae - 0%3B Bacteria - 15%3B Metazoa - 50%3B Fungi - 11%3B Plants - 76%3B Viruses - 3%3B Other Eukaryotes - 65 (source: NCBI BLink). | |||
| AT4G37250 | gene | 8.47 | 4:17527642 | Leucine-rich repeat protein kinase family protein%3B FUNCTIONS IN: protein serine/threonine kinase activity%2C kinase activity%2C ATP binding%3B INVOLVED IN: transmembrane receptor protein tyrosine kinase signaling pathway%2C protein amino acid phosphorylation%3B LOCATED IN: endomembrane system%3B EXPRESSED IN: 22 plant structures%3B EXPRESSED DURING: 13 growth stages%3B CONTAINS InterPro DOMAIN/s: Protein kinase%2C catalytic domain (InterPro:IPR000719)%2C Leucine-rich repeat-containing N-terminal domain%2C type 2 (InterPro:IPR013210)%2C Leucine-rich repeat (InterPro:IPR001611)%2C Serine/threonine-protein kinase-like domain (InterPro:IPR017442)%2C Protein kinase-like domain (InterPro:IPR011009)%3B BEST Arabidopsis thaliana protein match is: Leucine-rich repeat protein kinase family protein (TAIR:AT2G23300.1)%3B Has 127505 Blast hits to 87004 proteins in 3796 species: Archae - 113%3B Bacteria - 9564%3B Metazoa - 32249%3B Fungi - 5071%3B Plants - 68196%3B Viruses - 226%3B Other Eukaryotes - 12086 (source: NCBI BLink). | |||
| AT4G36770 | gene | 7.64 | 4:17330146 | UDP-Glycosyltransferase superfamily protein%3B FUNCTIONS IN: UDP-glycosyltransferase activity%2C transferase activity%2C transferring glycosyl groups%3B INVOLVED IN: metabolic process%3B LOCATED IN: cellular_component unknown%3B EXPRESSED IN: embryo%2C sepal%2C flower%2C pedicel%2C synergid%3B EXPRESSED DURING: 4 anthesis%2C C globular stage%3B CONTAINS InterPro DOMAIN/s: UDP-glucuronosyl/UDP-glucosyltransferase (InterPro:IPR002213)%3B BEST Arabidopsis thaliana protein match is: UDP-glucosyl transferase 72E1 (TAIR:AT3G50740.1)%3B Has 8229 Blast hits to 8179 proteins in 484 species: Archae - 0%3B Bacteria - 520%3B Metazoa - 2565%3B Fungi - 32%3B Plants - 4963%3B Viruses - 88%3B Other Eukaryotes - 61 (source: NCBI BLink). | |||
| AT4G36750 | gene | 6.96 | 4:17324519 | Quinone reductase family protein%3B FUNCTIONS IN: oxidoreductase activity%2C FMN binding%3B INVOLVED IN: negative regulation of transcription%3B LOCATED IN: plasma membrane%3B EXPRESSED IN: 24 plant structures%3B EXPRESSED DURING: 13 growth stages%3B CONTAINS InterPro DOMAIN/s: Flavoprotein WrbA (InterPro:IPR010089)%2C Flavodoxin/nitric oxide synthase (InterPro:IPR008254)%3B BEST Arabidopsis thaliana protein match is: flavodoxin-like quinone reductase 1 (TAIR:AT5G54500.1)%3B Has 3509 Blast hits to 3505 proteins in 1117 species: Archae - 64%3B Bacteria - 2674%3B Metazoa - 2%3B Fungi - 274%3B Plants - 203%3B Viruses - 1%3B Other Eukaryotes - 291 (source: NCBI BLink). | |||
| AT4G37580 | gene | HLS1 | 6.74 | 4:17658594 | involved in apical hook development. putative N-acetyltransferase | ||
| AT4G36791 | gene | 6.65 | 4:17338871 | This gene encodes a small protein and has either evidence of transcription or purifying selection. | |||
| AT4G36780 | gene | BEH2 | 6.62 | 4:17332269 | BES1/BZR1 homolog 2 (BEH2)%3B CONTAINS InterPro DOMAIN/s: BZR1%2C transcriptional repressor (InterPro:IPR008540)%3B BEST Arabidopsis thaliana protein match is: BES1/BZR1 homolog 1 (TAIR:AT3G50750.1)%3B Has 773 Blast hits to 298 proteins in 42 species: Archae - 2%3B Bacteria - 6%3B Metazoa - 9%3B Fungi - 12%3B Plants - 241%3B Viruses - 1%3B Other Eukaryotes - 502 (source: NCBI BLink). | ||
| AT4G37050 | gene | PLP4 | 5.85 | 4:17457135 | Patatin-related phospholipase A. Expressed in the floral gynaecium and is induced by abscisic acid (ABA) or phosphate deficiency in roots. | ||
| AT4G37640 | gene | ACA2 | 5.84 | 4:17682859 | Encodes a calmodulin-regulated Ca(2+)-pump located in the endoplasmic reticulum. Belongs to plant 2B ATPase's with an N-terminal autoinhibitor. | ||
| AT4G35830 | gene | ACO1 | 5.70 | 4:16972569 | Encodes an aconitase that can catalyze the conversion of citrate to isocitrate through a cis-aconitate intermediate%2C indicating that it may participate in the TCA cycle and other primary metabolic pathways. The protein is believed to accumulate in the mitochondria and the cytosol. It affects CSD2 (At2g28190 - a superoxide dismutase) transcript levels and may play a role in the response to oxidative stress. This enzyme can also specifically bind to the 5' UTR of CSD2 in vitro. | ||
| AT4G37100 | gene | 5.64 | 4:17479173 | Pyridoxal phosphate (PLP)-dependent transferases superfamily protein%3B FUNCTIONS IN: pyridoxal phosphate binding%2C catalytic activity%3B INVOLVED IN: biological_process unknown%3B LOCATED IN: plasma membrane%3B CONTAINS InterPro DOMAIN/s: Pyridoxal phosphate-dependent transferase%2C major domain (InterPro:IPR015424)%2C Pyridoxal phosphate-dependent transferase%2C major region%2C subdomain 1 (InterPro:IPR015421)%3B BEST Arabidopsis thaliana protein match is: Pyridoxal phosphate (PLP)-dependent transferases superfamily protein (TAIR:AT2G23520.1)%3B Has 644 Blast hits to 541 proteins in 145 species: Archae - 9%3B Bacteria - 25%3B Metazoa - 119%3B Fungi - 163%3B Plants - 272%3B Viruses - 0%3B Other Eukaryotes - 56 (source: NCBI BLink). | |||
| AT2G04030 | gene | CR88 | 5.10 | 2:01281754 | Encodes a chloroplast-targeted 90-kDa heat shock protein located in the stroma involved in red-light mediated deetiolation response and crucial for protein import into the chloroplast stroma. Mutants are resistant to chlorate%2C have elongated hypocotyls in light%2C and affect the expression of NR2%2C CAB%2C and RBCS but NOT NR1 and NiR. | ||
| GO | domain | Definition | P-value |
|---|---|---|---|
| aminopeptidase activity | molecular_function | "Catalysis of the hydrolysis of N-terminal amino acid residues from in a polypeptide chain." [GOC:jl, ISBN:0198506732] | 1.29e-02 |
| calcium-transporting ATPase activity | molecular_function | "Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: ATP + H2O + Ca2+(cis) = ADP + phosphate + Ca2+(trans)." [EC:3.6.3.8] | 1.29e-02 |
| plasma membrane | cellular_component | "The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins." [ISBN:0716731363] | 2.72e-02 |
| N-acetyltransferase activity | molecular_function | "Catalysis of the transfer of an acetyl group to a nitrogen atom on the acceptor molecule." [GOC:ai] | 1.48e-02 |
| indole glucosinolate metabolic process | biological_process | "The chemical reactions and pathways resulting in the formation of indole glucosinolates. Glucosinolates are sulfur-containing compounds that have a common structure linked to an R group derived from tryptophan; indoles are biologically active substances based on 2,3-benzopyrrole, formed during the catabolism of tryptophan." [http://www.onelook.com/] | 1.29e-02 |