|
Probe Labeling |
|||
|
|
Specific sequences in the restriction pattern on a Southern blot can then be identified by hybridization with a DNA probe. A probe is a piece of DNA identical (or very similar) to a sequence of interest. In order to locate a specific DNA sequence by hybridization, the probe is labeled with a reporter group. The Klenow fragment of E.coli DNA polymerase is used to make a
labeled probe. This so-called Klenow enzyme is a truncated
version of polymerase I: it lacks the 5’ to 3’ exonuclease activity. It
can synthesize DNA complementary to a DNA template in a 5’ to 3’
direction, provided it is given a primer with a free 3’-OH, and dNTP's.
A DNA fragment, often purified from agarose gel or a cleaned-up PCR
product, is chosen to act as a template in the polymerase
reaction. The duplex DNA is first denatured by boiling in the presence
of a mixture of all possible hexanucleotides (small, 6 nucleotides long
DNA molecules with a random sequence). These so-called random primers
anneal at positions of complernentarity along the single strands and act
as starting points for the Klenow polymerase to synthesize a
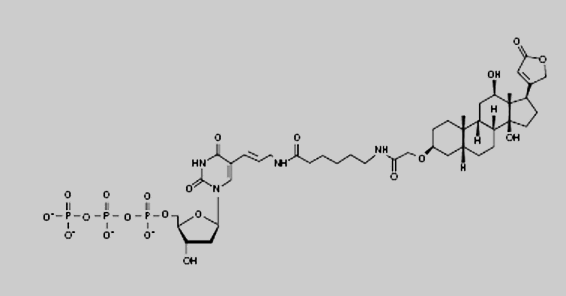
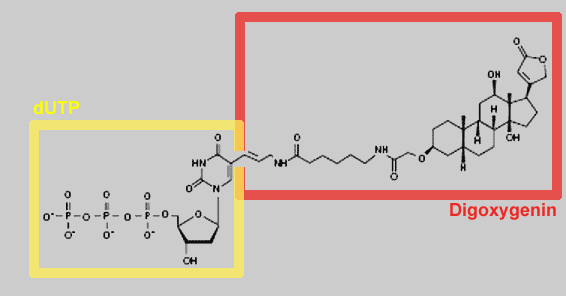
complementary strand. By using digoxygenin-labeled dNTPs in the
reaction mixture, the newly synthesized strands will be labeled with
digoxigenin. Eventually, a mixture of randomly sized labeled copy
fragments of both strands are present. note: a labeled probe can detect a band, even when it contains only a small part of the sequence present in that band! note:
DigoxigeninThe digoxygenin group is the label used in the Southern
hybridisation step in the Simple Cloning Lab.
Digoxigenin can be detected by antibodies, conjugated to the enzyme Alkaline Phosphatase. This enzyme can subsequently be used in a staining protocol, to visualise the localisation of labeled DNA.
|